|
Applications
& Technologies |
|
|
Summary Outline of Technology Info: A. Plasmids 1. Structure 2. Function 3. Recombinant DNA Procedure 4. Insulin Example B. Restriction Enzymes 1. Description 2. Methylation 3. Recognition Sequences 4. Production of gDNA 5. Blunt vs. Sticky Ends C. Oligonucleotides 1. Structure 2. Probes D. Vectors 1. Purpose 2. Size Restrictions 3. Plasmid vs. Lambda 4. Recombinant DNA Procedure E. Gel Electrophoresis 1. Purpose 2. Components 3. Analysis 4. Sample Gel Electrophoresis |
A. Plasmids
1. Structure
- self-replicating ring of DNA, containing 2-30 genes, found in bacterial
cells
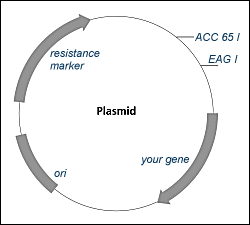
2. Function
- plasmids can be transferred quite easily from one bacterium through another
in a process called transformation; therefore, they are used frequently in
Recombinant DNA applications
** For more information on transformation, click on the Recombinant DNA
Applications link below.
3.
Recombinant DNA Procedure - steps to incorporate a foreign DNA sequence
into a plasmid
a. Cut open the plasmid
b. Splice other gene(s) into the plasmid
c. Use the recombined plasmid to make more copies of the inserted gene
and gene product

4. Insulin
Example - bacterial plasmid modified by the addition of the insulin gene
 a. Human insulin gene inserted into bacterial plasmid using the
Recombinant DNA Procedure outlined above.
a. Human insulin gene inserted into bacterial plasmid using the
Recombinant DNA Procedure outlined above.
b. Every time bacterium reproduces, more insulin gene (and insulin)
produced.
c. This insulin can be recovered and purified for use by diabetic
patients.
B. Restriction Enzymes
1.
Description - enzymes produced by bacteria in order to cut out foreign DNA
that may have been introduced by other bacteria
2.
Methylation - bacteria methylate, or add methyl groups (CH3) to
their own DNA so as to protect it from excision by restriction enzymes
3.
Recognition Sequences - sequences of DNA that are 4-8 base pairs in length;
these sequences must be located before a particular restriction enzyme is able
to initiate a cut
a. 90 different recognition sequences have been isolated from
approximately 200 different restriction enzymes
b. Examples:
HpaI
EcoRI
HindIII
5'
GTTAAC 3' 5'
GAATTC 3' 5' AAGCTT 3'
3' CAATTG
5' 3' CTTAAG
5' 3' TTCGAA 5'
4. Production
of gDNA - gDNA, otherwise known as genomic DNA, contains all of the DNA
fragments produced through the action of restriction enzymes
5. Blunt vs.
Sticky Ends - restriction enzymes can cut DNA sequences in these two
different methods
a. Blunt ends - the DNA fragments produced have even, straight-cut
ends
HpaI
5' GTT |||
AAC 3'
3' CAA |||
TTG 5'
b. Sticky
ends - the DNA fragments produced have uneven, overlapping ends; this
"cut and paste" technology allowed for the beginnings of Recombinant
DNA Applications
EcoRI
HindIII
5'
G |||
AATTC 3'
5' A |||
AGCTT 3'
3'
CTTAA |||
G
5'
3' TTCGA |||
A 5'
C. Oligonucleotides
1. Structure
- sequence of 12-20 DNA nucleotides produced in a laboratory machine
2. Probes
- oligonucleotides of a known sequence can be used to probe, or find, DNA to
which it is complementary; therefore, with whatever segment the probe binds to,
the DNA sequence can be determined
Probe -
AACCGGTT
DNA Sequence
- CCGATTCATTGGCCAATATTACCGGA
Probe + Sequence
- CCGATTCATTGGCCAATATTACCGGA
AACCGGTT
This allows the purple
segment of the
DNA sequence to be identified by base pairs due to complementary binding by the
probe.
D. Vectors
1. Purpose
- vectors are used to incorporate and produce gDNA or oligonucleotides in large
numbers (for instance it could be used to produce many copies of a particular
gene, similar to the insulin example above)
2. Size
Restrictions - most vectors can contain DNA inserts of up to 4,000 base
pairs in length
3. Plasmid
vs. Lambda
a. Plasmid - due to the small size of a plasmid itself, it can only
contain short DNA inserts of a couple thousand base pairs
b. Lambda bacteriophage - this larger vector can be used to
incorporate inserts of up to 20,000 base pairs; since Lambda is a bacterial
virus, every time the virus replicates, so does the insert
4.
Recombinant DNA Procedure - steps to follow in order to insert a DNA segment
into a vector
a. Cut vector with restriction enzyme
b. Cut insert with the same restriction enzyme
c. Use DNA Ligase to join the insert to the vector
d. Multiple copies of the vector are produced through the action of
bacteria (plasmid) or viruses (Lambda)
e. Cut the vector with the same restriction enzyme to remove the inserts.
f. Separate the inserts from the remaining gDNA using gel electrophoresis
(described below).
** To see a diagram of the Recombinant DNA Procedure described above, click on
the link below.
E. Gel Electrophoresis
1. Purpose
- technique that separates DNA fragments of different sizes after they have been
cut with restriction enzymes
2. Components
a. gDNA - segments of DNA that have previously been cut by
restriction enzymes
b. Agarose gel - substance with approximately the same consistency
as jello through which the gDNA segments will migrate based on their size
c. Electric Current - the gDNA segments are negatively charged due
to the presence of phosphate, thus they will migrate downward through the gel
towards the positive electric current
3. Analysis
- after the gDNA has migrated through the gel for several hours (through the
application of electricity), the gel must be stained and analyzed
a. gDNA migrates by size through the agarose gel
b. Large gDNA segments will remain close to the top of the gel
c. Small gDNA segments will travel to the bottom of the gel
d. Due to this separation pattern, segments of DNA (probes,
oligonucleotides) can be recovered by this technique if their nucleotide size is
known.
TOP - Negative
Electric Charge
---------- ---------- ---------- ---------- ---------- ---------- |
BOTTOM - Positive Electric Charge
Analysis of Gel by
gDNA Size (largest to smallest):
Purple
Blue
Orange
Green
Brown
Black
4. Sample Gel
Electrophoresis:

To test your knowledge about Recombinant DNA Techniques, click on the Technology Questions Link at the top of this page. After you answer the questions, be sure to check your responses by clicking on the Technology Answers Link.
|Home|
|Genetics| |Standardized
Exams|
|Ask a Question| |Related
Links| |Site Index|